A genome-wide association study of social genetic effects in Landrace pigs
Article information
Abstract
Objective
The genetic effects of an individual on the phenotypes of its social partners, such as its pen mates, are known as social genetic effects. This study aims to identify the candidate genes for social (pen-mates’) average daily gain (ADG) in pigs by using the genome-wide association approach.
Methods
Social ADG (sADG) was the average ADG of unrelated pen-mates (strangers). We used the phenotype data (16,802 records) after correcting for batch (week), sex, pen, number of strangers (1 to 7 pigs) in the pen, full-sib rate (0% to 80%) within pen, and age at the end of the test. A total of 1,041 pigs from Landrace breeds were genotyped using the Illumina PorcineSNP60 v2 BeadChip panel, which comprised 61,565 single nucleotide polymorphism (SNP) markers. After quality control, 909 individuals and 39,837 markers remained for sADG in genome-wide association study.
Results
We detected five new SNPs, all on chromosome 6, which have not been associated with social ADG or other growth traits to date. One SNP was inside the prostaglandin F2α receptor (PTGFR) gene, another SNP was located 22 kb upstream of gene interferon-induced protein 44 (IFI44), and the last three SNPs were between 161 kb and 191 kb upstream of the EGF latrophilin and seven transmembrane domain-containing protein 1 (ELTD1) gene. PTGFR, IFI44, and ELTD1 were never associated with social interaction and social genetic effects in any of the previous studies.
Conclusion
The identification of several genomic regions, and candidate genes associated with social genetic effects reported here, could contribute to a better understanding of the genetic basis of interaction traits for ADG. In conclusion, we suggest that the PTGFR, IFI44, and ELTD1 may be used as a molecular marker for sADG, although their functional effect was not defined yet. Thus, it will be of interest to execute association studies in those genes.
INTRODUCTION
The genetic effects of an individual may affect varieties of phenotypic traits. If these social effects on others are heritable they may affect response to selection, and thereby alter the outcome of both evolutionary processes in natural populations and artificial selection programs in agriculture [1,2]. Social genetic effects were observed in several livestock species, pig [3], chicken [4], and mink [5]. In pigs, individuals living in the same pen may show aggressive behaviors, such as tail biting, toward each other [6]. Such social interactions between pen-mates may affect their welfare and growth, such as average daily gain (ADG) considerably, both in negative and positive ways [7]. Previous studies evaluated the relationship between behavioral traits (i.e., aggressiveness and fearfulness) and the social genetic effects on ADG [6–8].
Since the very beginning of quantitative trait loci (QTL) mapping, about 14,479 QTLs for 592 different traits have been identified in the pig genome (PigQTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/SS/index). Most of the reported QTLs affect traits related to production and meat quality. For production traits, 562 QTLs have been identified for ADG. For exterior traits, 158 QTLs for behavior (February 2016). In particular, QTL associated with the time spent socializing were found on chromosome numbers 6, 7, 11, 12, and 13. QTL for individual ADG has been reported on almost all chromosomes in pig [9], suggesting that production efficiency, such as ADG and feed efficiency is a multiple-loci trait. Unlike individual ADG, little is known about the effects of candidate genes on ADG of social mates. The purpose of this study was to identify candidate genes for ADG of social mates in pig using the genome-wide association approach in a Landrace population.
MATERIALS AND METHODS
Animals and phenotypes
The phenotypic dataset on ADG of animals was obtained through performance tests of Landrace pigs (n = 20,130) between 2005 and 2015 in South Korea (Landrace Sunjin, Republic of Korea, http://dad.fao.org/). These pigs were fed ad libitum, and water was constantly accessible through nipple drinkers. Performance evaluation for individual ADG (iADG) was initiated when the pigs were 29.9±3.5 kg in live weight and ended when the animals reached 89.9±8.2 kg. Social ADG (sADG) was the average ADG of unrelated pen-mates (strangers). The sADG dataset consisted of 16,802 records, excluding the pens with only full-sib mates. We used the phenotype data after correcting for batch (week), sex, pen, number of strangers (1 to 7 pigs) in the pen, full-sib rate (0% to 80%) within pen, and age at the end of the test.
Genotyping and quality control
Genomic DNA of Landrace pigs was extracted from blood samples using DNA isolation kit (Promega, wizard genomic DNA purification kit, Madison, WI, USA) according to manufacturer’s instruction. A total of 1,041 pigs from Landrace breeds were genotyped using the Illumina PorcineSNP60 v2 BeadChip panel, which comprised 61,565 single nucleotide polymorphism (SNP) markers.
Quality control was carried out using PLINK software. SNP markers were removed if they had genotype missing rate >0.1 (GENO), minor allele frequencies (MAF) <0.05, and Hardy–Weinberg Equilibrium <0.0001. We selected individuals that had call rate >0.90 (MIND). Finally, 39,837 markers and 1,038 subjects (909 subjects for sADG) passed the quality control and were used in the following analysis.
Genome-wide association study (GCTA implementation of MLMi and MLMe)
The genome-wide association study (GWAS) and heritability estimate were conducted using a mixed linear model (MLM) based association analysis by estimating the genetic relationship matrix (GRM) implemented in GCTA software. We implemented the MLM based association analysis by including (MLMi) and excluding (MLMe) the candidate SNPs [10].
The phenotype y for MLMi using MLMA option is modeled as:
where c is a vector of fixed covariates (including the affine term) with corresponding coefficient matrix K; g is a vector of genetic effects with
RESULTS AND DISCUSSION
The final dataset contained genotypes from a total of 1,038 pigs (909 pigs for sADG) for GWAS. The descriptive statistics of the observed and adjusted phenotype values are displayed in Table 1. The heritability was 0.43 and 0.14 in iADG and sADG, respectively (Table 2). This result indicates that, in addition to being influenced by the environment, iADG and sADG are affected by an additive genetic component, a finding justifying the execution of GWAS.

Estimates of variances explained by SNPs for individual average daily gain (iADG) and social average daily gain (sADG) of Landrace nucleus herd
A Q–Q plot comparing the distribution of observed chi-square statistics with the distribution of those expected under the null hypothesis is shown in Figure 1. For the MLMi method, the genomic inflation factors for iADG and sADG are 1.02 and 1.00, respectively. However, MLMe resulted in a high genomic inflation factor (1.28) for iADG, but not for sADG (1.07). Therefore, after checking the population structure (Figure 2), we only described the result of MLMi method for iADG. A Manhattan plot showing the GWA results is presented in Figure 3. There are a significant number of markers that surpass the suggestive significance level (FDR≤0.20) in MLMe, and a summary of these SNPs, their map locations, and their p-values and FDR are reported in Table 3. There is no significant SNPs for iADG. A total of 5 SNPs for sADG, which passed the suggestive significance level (FDR≤0.20), were identified on Sus scrofa chromosome (SSC) 6 and had the same MAF (0.19) and FDR (0.15). INRA0022326 SNP lies within the prostaglandin F2α receptor (PTGFR) gene and this SNP is placed 124,909,803 bp on SSC6. Prostaglandin F2α is involved in several reproductive processes, including the physiological changes associated with farrowing. Prostaglandin F2α is considered as principal luteolysin in domestic animals, which acts on the target cells via a specific plasma membrane-associate receptor (PTGFR [11]). This receptor is already abundant on the surface of porcine luteal cells during the early luteal phase [12]. On the other hand, repeated administration of prostaglandin F2α on day 5 of the estrous cycle does promote luteolysis in pigs [13]. Despite previous studies, the molecular mechanism of luteolytic sensitivity acquisition in porcine corpora lutealis not fully understood. The PTGFR is known to affect sexual behavior, which can also be influenced by the social environment in pig [14]. They reported that the social environment after puberty affects the sexual behavior and the social restriction during rearing causes the depression in sexual behavior. Therefore, the sexual behavior is connected with the social interaction. Various behaviors altered through selection of social genetic effects appear to reflect an internal state rather than solely social interactions [15]. ASGA0029469 SNP is not inside the gene, but is located at 22,198 bp upstream of the interferon-induced protein 44 (IFI44). IFI44 was first reported in the liver of chimpanzees infected with hepatitis C [16]. The IFI44 gene is an inducible interferon-specific gene. Its expression is up-regulated in the presence of interferon in cell cultures and it may be involved in anti-hepatitis C virus activity [17]. However, its function remains unclear. As proposed by Hallen et al [18], IFI44 may induce a cellular GTP depletion that abolishes extracellular signal-regulated kinase signalling and results in cell cycle arrest. Green et al [19] reported a homologous gene in the Sydney rock oyster, S. glomerata. In humans, IFI44 localizes to the nuclei and suppresses the HIV-1 LTR promoter [20] and is related to chronic neurodegenerative diseases, such as Alzheimer’s disease [21]. Ueyama et al [22] reported that p44/p42 mitogen-activated protein kinase is related to emotional stress of rats. Supporting the association between IF144 and emotional stress [22], pigs with high social genetic effects were better able to manage stressful situations and were less fearful because of the apathy of both the animals and the situations [6–8]. In addition, the emotional decision in pigs is similar to humans. Asher et al [23] reported that the judgments and decisions of a pig are governed by their mood and their personality type. Further studies for verification of genes related to social genetic effects in emotion change per situation and in coping with stress are required. LGA0114621, MARC0099561, and ALGA0119254 SNPs are located 160,543, 176,089, and 190,899 bp upstream of EGF latrophilin and seven transmembrane domain-containing protein 1 (ELTD1), respectively. ELTD1 is not well characterized. Based upon its sequence, ELTD1 is a member of the secretin family of G-protein-coupled peptide hormone receptors and belongs to the epidermal growth factor-seven-transmembrane (EGF-TM7) subfamiliy. Structurally, it contains a large extracellular domain with EGF-like repeats, a seven-transmembrane domain, and a short cytoplasmic tail. ELTD1 was first identified to be developmentally regulated in rat fetal and postnatal cardiomyocytes [24]. ELTD1, an orphan G-protein-coupled receptor (GPCR) belonging to the adhesion GPCR family, was recently linked to angiogenesis in humans [25]. Up-regulation of the ELTD1 gene was observed in previous study describing the genetic predisposition for obesity in both humans and pigs [26]. Agrawal et al [27] reported that ELTD1 was an important candidate gene for genetic risk to cannabis use disorders in neuropeptide signaling and signal transduction pathways.
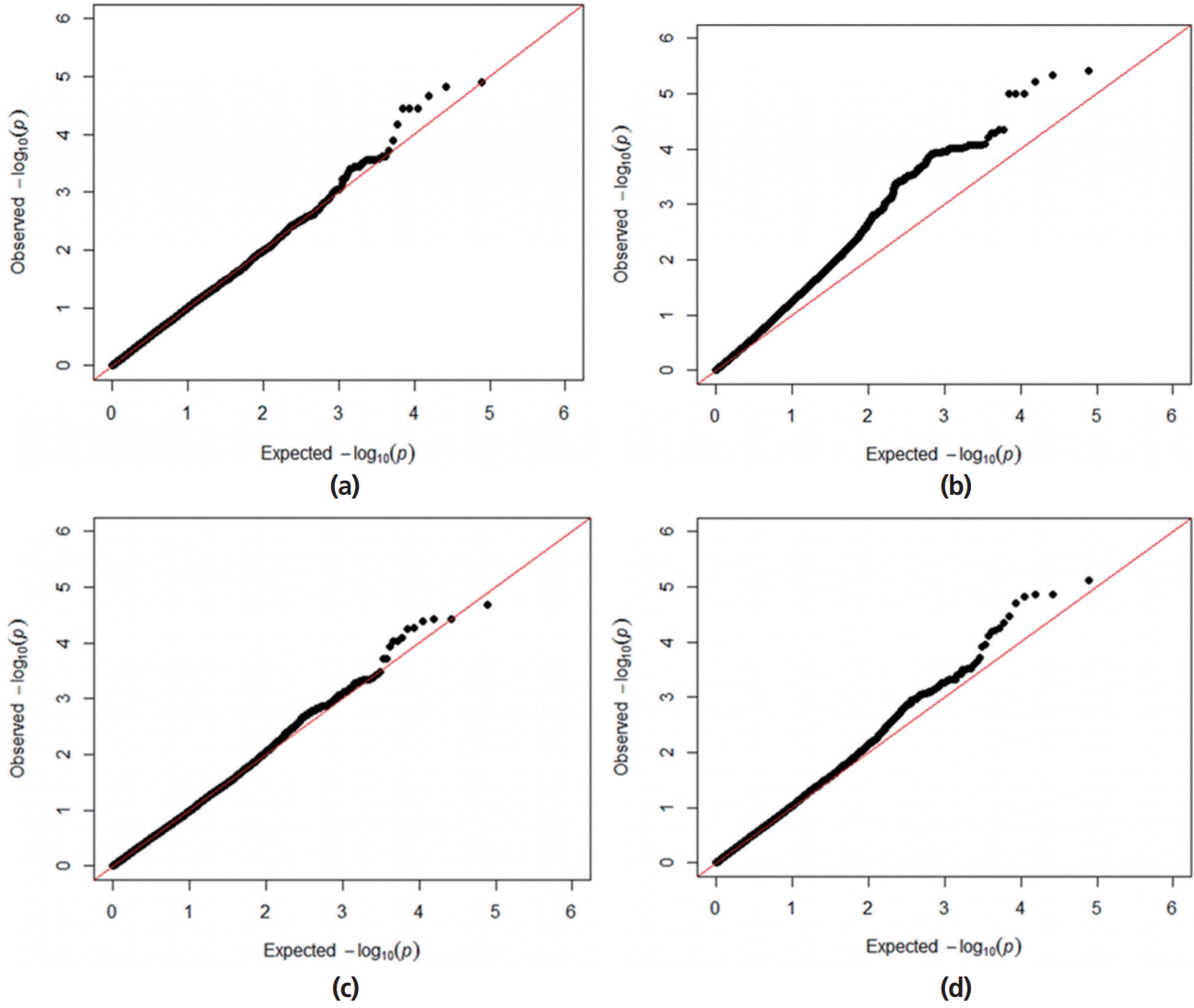
Quantile-quantile plots for average daily gain (ADG) using the mixed-model association methods with different genomic inflation factor (lambda) in a Landrace nucleus herd. (a) Quantile-quantile plot with lambda of 1.02 for individual average daily gain (iADG) with candidate marker included mixed linear model (MLMi). (b) Quantile-quantile plot with lambda 0f 1.28 for iADG with candidate marker excluded mixed linear model (MLMe). (c) Quantile-quantile plot with lambda of 1.00 for social average daily gain (sADG) with candidate marker MLMi. (d) Quantile-quantile plot with lambda of 1.07 for sADG with candidate marker MLMe.

Manhattan plots for average daily gain (ADG) using the mixed-model association methods in a Landrace nucleus herd. The horizontal red line indicates the significance thresholds (false discovery rate ≤0.20). (a) Manhattan plot for individual average daily gain (iADG) with candidate marker included mixed linear model (MLMi). (b) Manhattan plot for iADG with candidate marker excluded mixed linear model (MLMe). (c) Manhattan plot for social average daily gain (sADG) with candidate marker MLMi. (d) Manhattan plot for sADG with candidate marker MLMe.
In pigs, the magnitude of social genetic effects has been estimated for ADG [3,28] and total heritable variance expressed relative to phenotypic variance clearly exceed the usual range of heritability for ADG and feed intake [3]. However, there have been no previous association studies with social genetic effects for ADG. As potentially affecting molecule, androstenone influences social behaviors, such as tail biting and aggression [29]. Therefore, it is important to consider androstenone with the social genetic effects as a social behavior feature. Duijvesteijn et al [30] detected new genes on chromosome 6 from the pathways of the synthesis and metabolism of androstenone such as α-chain of the luteinizing hormone (LHA), β-chain of the luteinizing hormone (LHB) and hydroxysteroid-dehydrogenases (HSD17B14). In current study, although SNPs for social genetic effects differ from them for androstenone, significant markers (5 SNPs) for social genetic effects located intensively on chromosome 6. The LHB gene which induces steroid synthesis in the Leydig cells of the testis at onset of puberty maps to this area on chromosome 6. Thus, further research is needed to identify the SNPs on chromosome 6 that are associated with social genetic effect and reproductive feature.
The results obtained from GWAS for social ADG in the Landrace breed identified new genomic regions and genes associated with social ADG in the porcine genome. PTGFR, IFI44, and ELTD1 have never been reported with social interaction and social genetic effect in any of the previous studies. Identification of several genomic regions and putative positional genes associated with social genetic effects, as reported here, should contribute to a better understanding of the genetic basis of interaction traits.
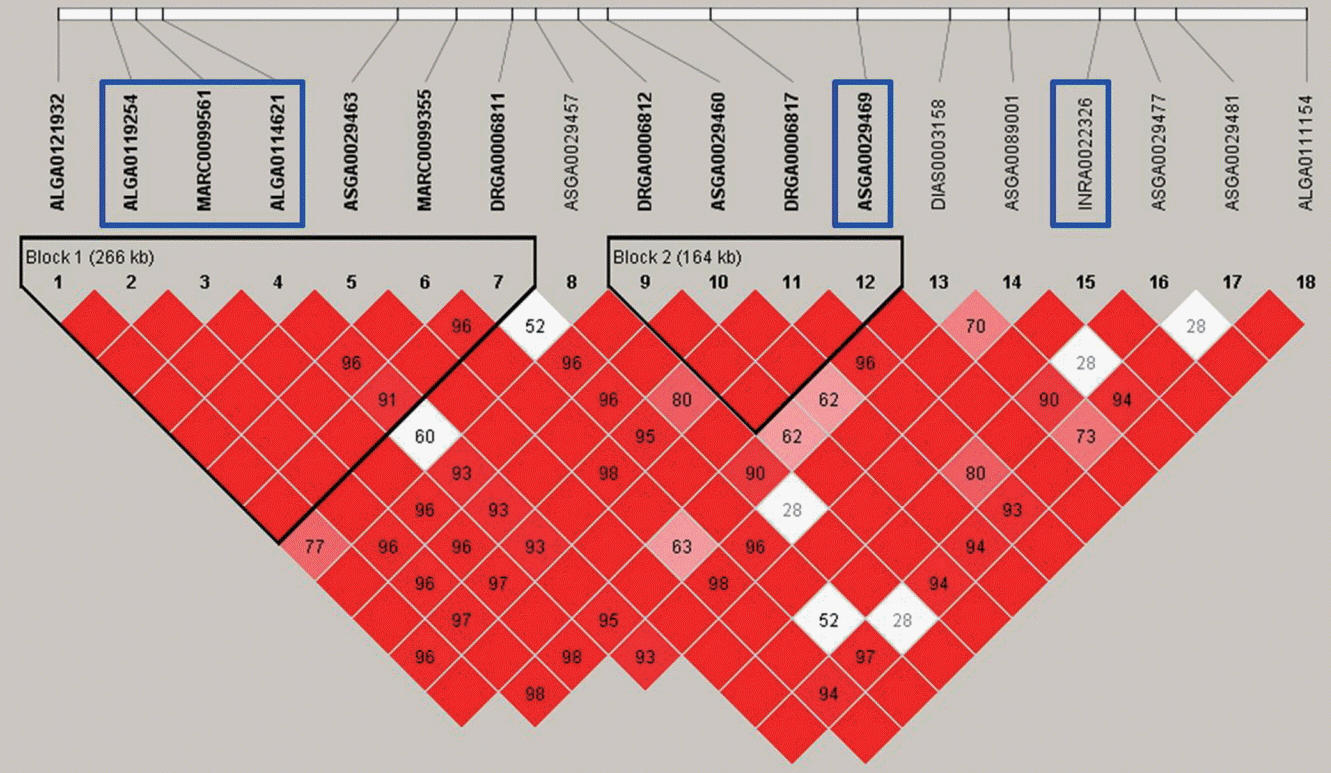
Linkage disequilibrium (LD) plot of the 266 kb region where markers ALGA0119254, MARC0099561, and ALGA0114621 are localized for social average daily gain (sADG). The coefficient of LD (r2) between significant single nucleotide polymorphisms (SNPs) was calculated within 500 kb chromosomal distance and the complete linkage (r2 = 1) was considered to a haplotype block represented by red diamonds.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
ACKNOWLEDGMENTS
The design of the study was financially supported by the Rural Development Administration in Republic of Korea (PJ00997101). The collection of data and analysis were funded by 2018 the RDA Fellowship Program of National Institute of Animal Science, Rural Development Administration, Republic of Korea.



