INTRODUCTION
Identification of genetic markers in relation to phenotypic differences is important for chicken breed identification. Enormous progress with MC1R gene polymorphisms has been achieved in relation to coat color variations in vertebrate animals (Robins et al., 1993; Jackson et al., 1994; Kerje et al., 2003). In the chicken, the MC1R gene plays a key role in the regulation of eumelanin (black/brown) and pheomelanin (red/yellow) feather pigmentation (Robbins et al., 1993; Jackson, 1997). The chicken MC1R gene is located on chromosome 11 and encodes a 314 amino acid protein in only one exon (Takeuchi et al., 1996; Kerje et al., 2003). Mutations in the MC1R gene have been associated with coat color variations in horse, cattle, fox, pig, sheep, dog, quail, guinea fowl and chicken (Klungland et al., 1995; Marklund et al., 1996; Vage et al., 1997; 1999; Kijas et al., 1998, 2001; Newton et al., 2000; Kerje et al., 2003; Nadeau et al., 2006; Vidal et al., 2010). Previous study was investigated for the relationships of the plumage colors with TYR and MC1R genes in Korean chickens (Heo et al., 2011). Interestingly, the MC1R gene has been sequenced in a variety of avian species and has been identified as a possible signature of sexual selection among sexually dimorphic birds (Nadeau et al., 2007). The melanogenesis pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) includes a few expressed genes which containing tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1) and dopachrome tautomerase (DCT). Furthermore, preliminary data indicate that the agouti-related protein (AGRP) gene controls melanin synthesis and processing of endogenous antagonist of the melanocortin system (Nadeau et al., 2007). Also, the MC1R gene is especially known for its association with melanic polymorphisms in vertebrate species (Hoekstra et al., 2006).
Different colors in Korean native chickens have fundamental importance for identifying breeds. In 1994, the Korean government started a conservation project and collected native chickens on the basis of phenotypic appearance, mostly based on feather colors. The current Korean native chicken breeds have five different feather colors (black, red, yellow, gray and white). Therefore, the genetic markers responsible for feather colors are good candidates for breed identification, especially for the Korean native chickens. In this study, the MC1R gene variations were investigated for their possible use in breed identification of differently colored Korean native chicken breeds.
MATERIAL AND METHODS
Sampling and DNA extraction
Five chicken breeds consisting of total 150 individuals (30 samples in each breed) were used in this study. They were four Korean native chicken (KNC) breeds including KNC black (KB), KNC yellow (KY), KNC red (KR) and black Silky (O). Also, White Leghorn (W) was included as a control breed having white plumage color (Table 1). Blood samples for all individuals were collected from NIAS in Korea. Genomic DNAs were extracted using Prime Prep⢠Genomic DNA Isolation Kit (GeNetBio, Korea) according to the manufacturer's instructions.
PCR amplification and DNA purification
Primers were designed from National Center for Biotechnology Information (NCBI) reference data (GenBank accession no. D78272) using Primer3 program (v. 0.4.0). The forward (5â˛-GCTTTGTAGGTGCTGCAG TTGTG-3â˛) and reverse primer (5â˛-CCATCCATCCTC CTGTCTGT-3â˛) were used to PCR amplify a 1,050 bp fragment in the MC1R gene. The PCR reactions included approximately 100 ng of genomic DNA, 2.5 Îźl of 10Ă buffer (Tris-HCl, pH 9.0), PCR enhancers, (NH4)2SO4, 20 mM MgCl2), 2.0 Îźl of 10 mM dNTPs mixture (2.5 mM each of dATP, dCTP, dGTP and dTTP), 1 Îźl of 10 pmol of each primer and 1 U HS Prime Taq (GeNet Bio, Korea) in a 25 Îźl reaction volume. PCR was performed in a My-Genie 96 Thermal Block (Bioneer) with an initial denaturation step at 94°C for 10 min followed by 35 cycles of 30 s at 94°C, 30 s at 61°C, 40 s at 72°C and a final extension step at 72°C for 10 min. The PCR products were electrophoresed on 1.5% agarose gels stained with ethidium bromide, and DNA fragments were visualized under ultraviolet light. Purification of PCR products was performed using an AccuPrepÂŽ PCR purification kit (Bioneer) according to the manufacturerâs instructions. Purified PCR products were also confirmed by using agarose gels for sequencing.
Sequencing and genotyping
All the purified PCR products were sequenced by Genotech (www.genotech.co.kr). For restriction fragment length polymorphism (RFLP) genotyping, three selected SNPs at nucleotide position 69, 376 and 427 were used. Based on the sequence alignment, KR, KY and W chicken breeds have C SNP in 69 nucleotide position. Also, W chicken breed has A SNP in 376 nucleotide position. However, KR and KY chicken breeds have G SNP in 427 nucleotide position. Therefore, using these three SNPs, Korean native chicken breeds were well characterized. The amplified MC1R PCR products were digested with BsrDI, Hpy99I and BtgZI restriction enzymes, respectively. The RFLP reaction consisted of 10Ă buffer, 3 to 5 units of each enzyme and 15 Îźl of PCR products in a total volume of 20 Îźl. The obtained RFLP fragments were separated on 3% agarose gels stained with ethidium bromide.
Analysis
The chicken MC1R gene nucleotide sequences were aligned using the ClustalW program (Thompson et al., 1994) and saved in the Bioedit format. Nucleotide replacement export data from the MC1R gene were carried out for polymorphisms and constructed phylogenetic analysis using MEGA software version 4.0.2 (Kumar et al., 2008). Genotype and allele frequencies of the five breeds were calculated based on the PCR-RFLP results. Also, haplotype frequencies were investigated using the algorithm from the Haploview program (Haploview, USA).
RESULTS AND DISCUSSION
SNP identification and phylogenetic analysis
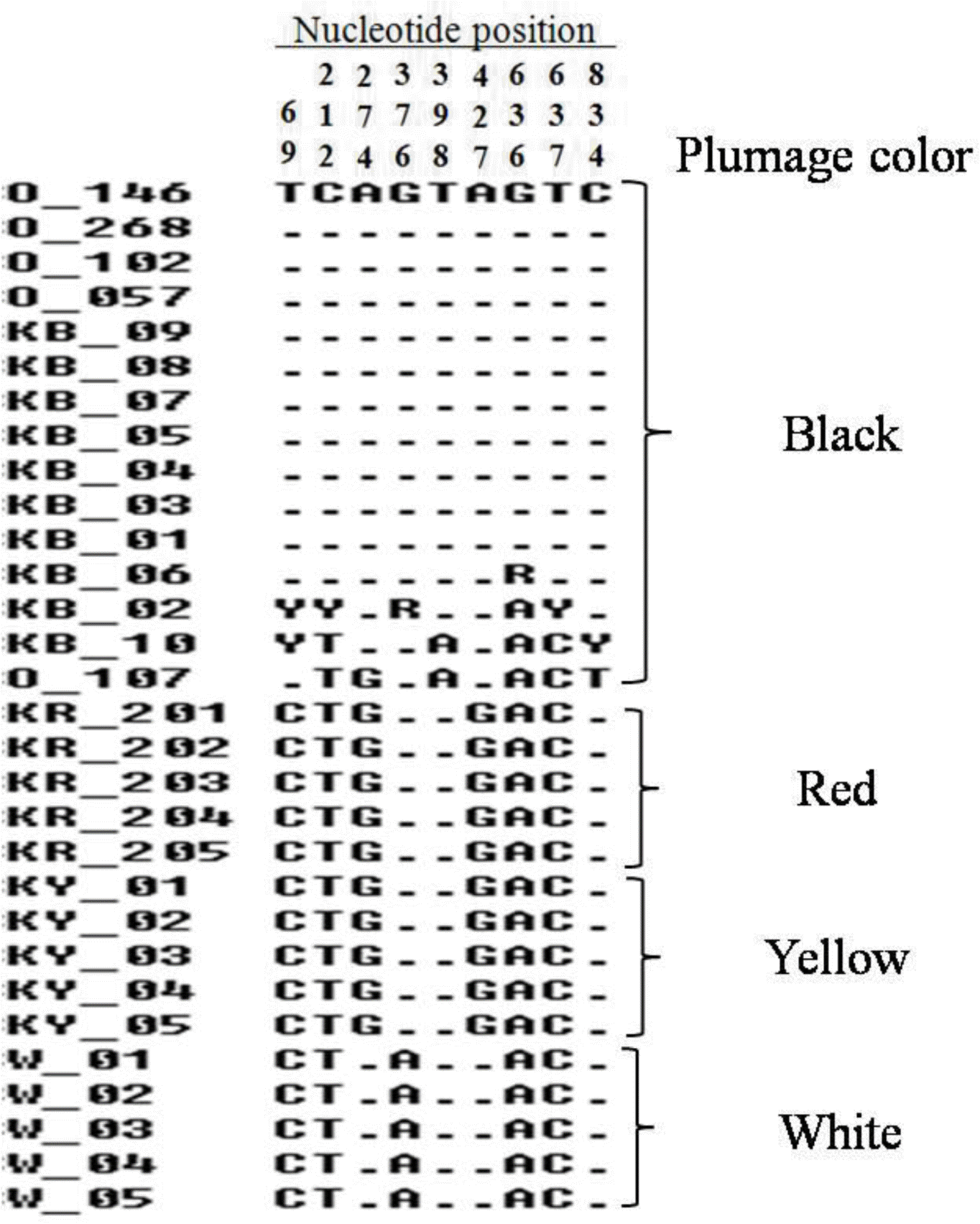
Four Korean native chicken breeds and White Leghorn as a control breed were used for the identification of MC1R gene polymorphisms. A total of 30 individuals were initially sequenced and nine polymorphisms were obtained. Of these, three polymorphisms (nucleotide positions at 69, 636 and 834) were the synonymous mutations, while six polymorphisms (nucleotide positions at 212, 274, 376, 398, 427 and 637) were nonsynonymous mutations (Figure 1). When we look at the breeds having black plumage colors, 11 out of 15 sequences are identical and four sequences have heterozygous SNPs (Figure 1). These four sequences are from Korean native chicken black (animal ID: KB_02, KB_06 and KB_10) and black Silky (Animal ID: O_107). Previously, plumage color dominance allele series in the MC1R gene have been related with pigmentation (Ling et al., 2003). Therefore, the mutations in this gene can affect eumelanin deposition, which gives the darker pigmentation. On the other hand, the mutation of this gene also can suppress the activity of the MC1R gene, indirectly leading to more pheomelanin pigmentation (Guo et al., 2010). Even though, those different polymorphisms in black feathered breeds have been expressed to black pigmentation phenotype. Previously, 34 SNPs of the MC1R gene have been investigated in several chicken populations (Kerje et al., 2003; Guo et al., 2010; Tixier-Biochard, 2011). Eight causative nonsynonymous mutations were suggested for the relationship with eumelanin and pheomelanin pigments on plumage colors (Kerje et al., 2003; Ling et al., 2003). In case of our study, six nonsynonymous mutations were observed in Korean native chickens. Previous study has been reported that six nonsynonymous mutations (Met71Thr, Glu92Lys, Ala126Ile, Thr143Ala, Cys213Arg and His215Pro) were highly associated with eumelanin and pheomelanin pigmentation. These mutations were associated in chicken plumage colors (Kerje et al., 2003; Ling et al., 2003; Guo et al., 2010). In this study, the investigation of MC1R gene polymorphisms in Korean native chicken confirms the previous finding for the plumage color discrimination using MC1R gene polymorphisms.
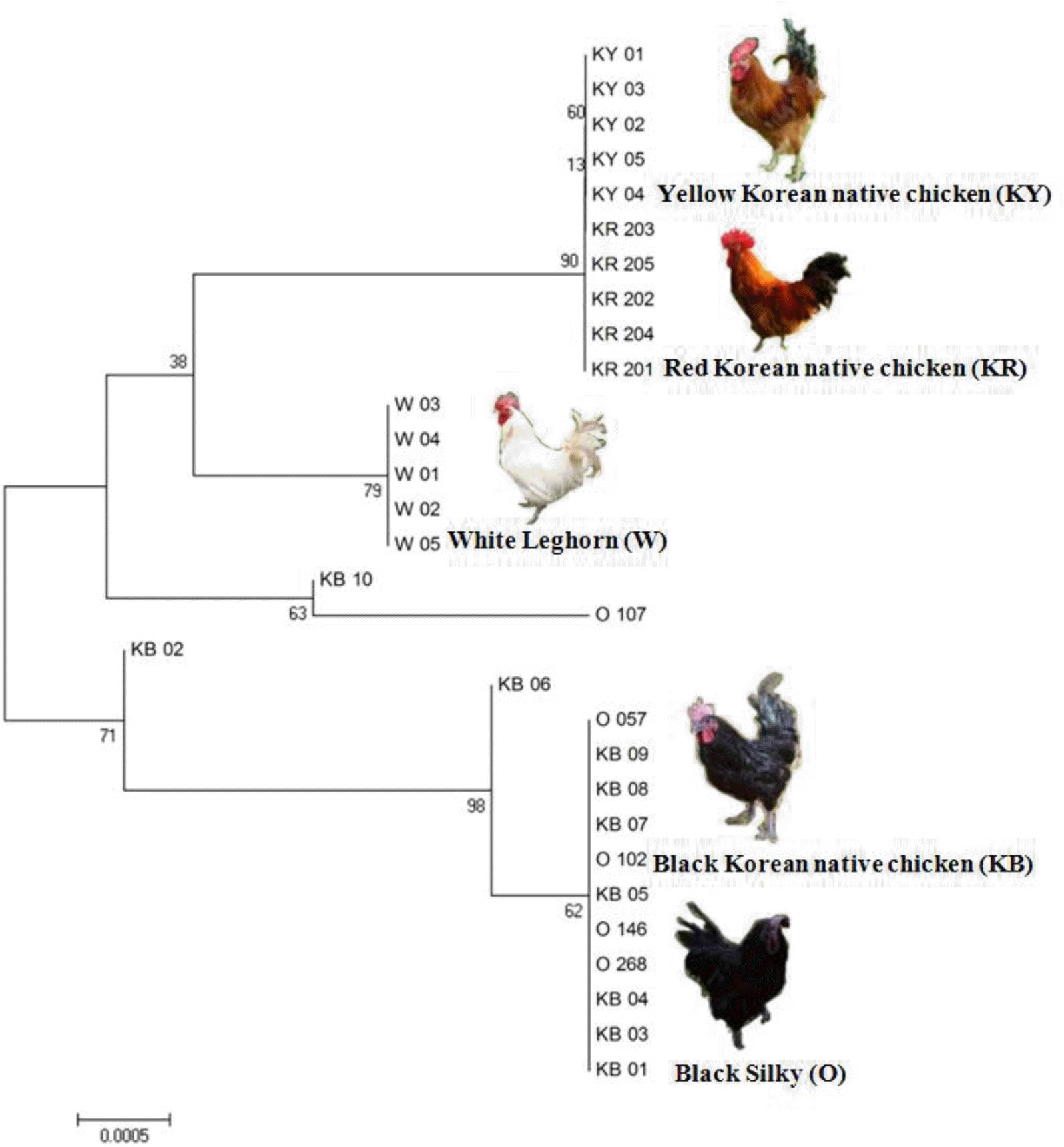
Based on the MC1R gene sequencing of 30 individuals in five chicken breeds, a neighbor-joining (NJ) phylogenetic tree was constructed. The results demonstrated the clear separation of the chicken breeds along with plumage color variations (Figure 2). Two chicken breeds (Black Korean native chicken and Black Silky) were colored animals responsible for eumelanin (black/brown) and two other breeds (Red Korean native chicken and Yellow Korean native chicken) were pheomelanin (red/yellow). Also, the control White Leghorn breed was responsible for albino. Interestingly, the phylogenetic analysis revealed that eumelanin containing chicken breeds are more ancient than pheomelanin and albino containing breeds. Based on the previous results, plumage colors do not depend on only one mutation. Several nucleotide substitutions in the chicken MC1R gene are associated with different phenotypes, from the dominant extended black to the recessive yellow (Takeuchi et al., 1996; Kerje et al., 2003; Ling et al., 2003).
Genotype and allele frequency
On the basis of three selected SNPs, genotype and allele frequencies were calculated in five chicken breeds (Table 2). Genotype distribution for the 69T>C SNP in each breed indicated that TT genotypes were clearly shown in KB (56.7%) and O (80%) chicken breeds, while the CC genotype was represented in KB (3.3%), KY (80%), KR (66.7%) and W (100%) chicken breeds. In this SNP, black feathered chicken breeds have high frequencies of the T allele (76.7% KB and 90% O). In case of the breeds having other feather colors (83.3% red, 90% yellow and 100% white), the C allele was observed in high frequency. For the 376G>A SNP, the GG genotypes were identified in KB, O, KY and KR breeds, whereas the GG genotype did not appear in the White Leghorn breed. In contrast, the AA genotype was observed only in White Leghorn. In contrast, KB (86.7%), O (100%), KY (100%) and KR (100%) chicken breeds displayed high allele frequencies in G allele, while A allele frequencies were high in the white feathered breed. In the case of 427A>G SNP, high frequencies of the GG genotype were observed only in red (63.3%) and yellow (66.7%) feathered breeds, while the black and white feathered chicken breeds displayed predominant AA genotypes. Therefore, only the A allele was observed in KB, O and W breeds, while red and yellow feathered breeds contained the G allele with high frequencies. Previously, these SNPs have been associated in plumage colors in Chinese chickens (Guo et al., 2010). The interesting phenomenon found in this study was the specific MC1R genotype distributions in these breeds, which was strongly associated with the molecular configuration of plumage color pigmentation.
Haplotype frequency
The three selected SNPs were genotyped in five chicken breeds and haplotype frequencies were calculated (Table 3). Five haplotypes were identified. TGA haplotype displayed high frequencies in KB (76.7%) and O (90%) breeds, low frequencies in KY (10%) and KR (15%) breeds and no TGA haplotype was observed in the W breed. On the other hand, the CGG haplotype appeared only in the KY (66.7%) and KR (61.7%) chicken breeds. Also, the CAA haplotype was observed in the W breed (91.7%) with high frequencies and in low frequencies (13.3%) in the KB breed. All five examined breeds possessed the CGA haplotype. Also, a low frequency of TGG haplotype was observed only in the KR breed (1.7%). Based on the estimation of haplotypes from three SNPs, three haplotypes (TGA, CAA and CGA) are representing the black chicken breeds. Also, CAA and CGA haplotypes were identified in White Leghorn, whereas these haplotypes in black chicken breed as a low frequencies. On the other hand, pheomelanin (yellow/red) chicken breed was obtained four haplotypes (TGA, CGG, CGA and TGG), where CGG haplotype is unique.
This study shows the MC1R gene polymorphisms and their relationships with plumage color variation in Korean chicken breeds. To maintain the valuable native chicken breeds, an appropriate conservation breeding program will ultimately be needed. Based on the results presented here, molecular markers in MC1R gene provide some directions for the identification of plumage color-based breed identification in Korean chicken breeds. To improve the power of breed discrimination, more molecular markers need to be studied.




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print







