1. Feil R, Khosla S. Genomic imprinting in mammals: an interplay between chromatin and DNA methylation? Trends Genet 1999; 15:431–5.


2. Lesch BJ, Silber SJ, McCarrey JR, Page DC. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet 2016; 48:888–94.


3. Waclawska A, Kurpisz M. Key functional genes of spermatogenesis identified by microarray analysis. Syst Biol Reprod Med 2012; 58:229–35.


5. Zhang B, Zheng H, Huang B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016; 537:7621553–7.


6. Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8:532–8.


9. Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 2010; 339:240–9.


11. Li A, Figueroa S, Jiang T-X, et al. Diverse feather shape evolution enabled by coupling anisotropic signalling modules with self-organizing branching programme. Nat Commun 2017; 8:ncomms14139

14. Chokeshaiusaha K, Thanawongnuwech R, Puthier D, Nguyen C. Inspection of C-type lectin superfamily expression profile in chicken and mouse dendritic cells. Thai J Vet Med 2016; 46:443–53.

16. Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21.


19. Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc 2012; 7:1728–40.


21. Ross-Innes CS, Stark R, Teschendorff AE, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012; 184:7381389–93.




 , peaks shared among all samples with either K4 or K27 trimethylation (All_shared_K4_and_K27);
, peaks shared among all samples with either K4 or K27 trimethylation (All_shared_K4_and_K27);
 , peaks presented only among samples with K4 trimethylation (All_shared_K4_but_not_K27);
, peaks presented only among samples with K4 trimethylation (All_shared_K4_but_not_K27);
 , peaks presented only among samples with K27 trimethylation (All_shared_K27_but_not_K4);
, peaks presented only among samples with K27 trimethylation (All_shared_K27_but_not_K4);
 , peaks presented only in Lat_K4 (Unique_Lat_K4);
, peaks presented only in Lat_K4 (Unique_Lat_K4);
 , peaks presented only in Med_K4 (Unique_Med_K4);
, peaks presented only in Med_K4 (Unique_Med_K4);
 , peaks presented only in PS_K4 (Unique_PS_K4);
, peaks presented only in PS_K4 (Unique_PS_K4);
 , peaks presented only in RS_K4 (Unique_RS_K4);
, peaks presented only in RS_K4 (Unique_RS_K4);
 , peaks presented only in PS_K27 (Unique_PS_K27);
, peaks presented only in PS_K27 (Unique_PS_K27);
 , peaks presented only in RS_K27 (Unique_RS_K27), and
, peaks presented only in RS_K27 (Unique_RS_K27), and
 , other peaks not in the previously mentioned groups. Distribution of precipitated DNA fragments acquired from sample ChIP-seq data according to chicken gene transcription start site was also demonstrated (B). Aggregation of fragments within 0–1 kilobases (
, other peaks not in the previously mentioned groups. Distribution of precipitated DNA fragments acquired from sample ChIP-seq data according to chicken gene transcription start site was also demonstrated (B). Aggregation of fragments within 0–1 kilobases (
 ) and 1–3 kilobases (
) and 1–3 kilobases (
 ) are presented.
) are presented.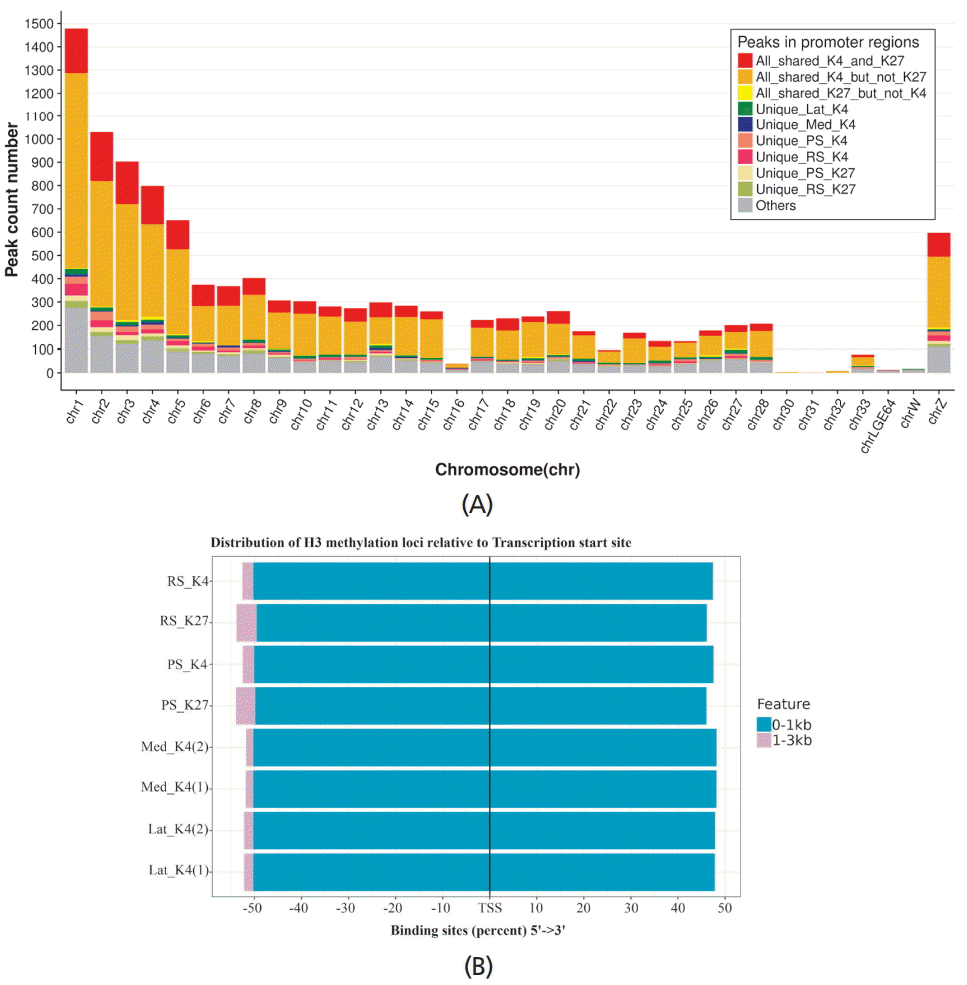
 ) and orange (
) and orange (
 ) color, accordingly. The horizontal leftmost bar graph (olive green color,
) color, accordingly. The horizontal leftmost bar graph (olive green color,
 ) show to total number of peaks presented in each cell type.
) show to total number of peaks presented in each cell type.
